Cytochrome P450 Enzymes; CYPs
My basic research program focuses on the cytochrome P450 superfamily of proteins with an emphasis on their evolution and function in aquatic species such as fish. Cytochrome P450 enzymes, or CYPs, are heme proteins critical for generation of major biological signaling molecules (e.g. steroid hormones) and for the detoxification of xenobiotics (e.g. drugs, environmental contaminants). CYPs are a key component of the defensome – the genes that aid in protection and defense from toxic compounds. Vertebrate species have 50-100 CYP genes in their genome but the function of many of these genes in non-mammalian species is unclear. We have a primary interest in understanding the evolution and function of CYPs in aquatic organisms.
Our projects involve genome annotation of CYP sequences, phylogenetic studies of CYP families, protein expression and functional testing of CYPs. This research raises fundamental questions about CYP protein function and attracts students with strong interests in protein evolution, bioinformatics, molecular biology and biochemistry.
Bioinformatics approaches are used in genome annotations of CYP genes and phylogenetic studies that raise functional hypotheses regarding novel CYP sequences. With each new genome completed, an array of CYP sequences are identified for which functional knowledge is lacking. Our basic science research is directly aimed at uncovering the function of these novel genes and understanding the capacity of CYP systems in aquatic species.
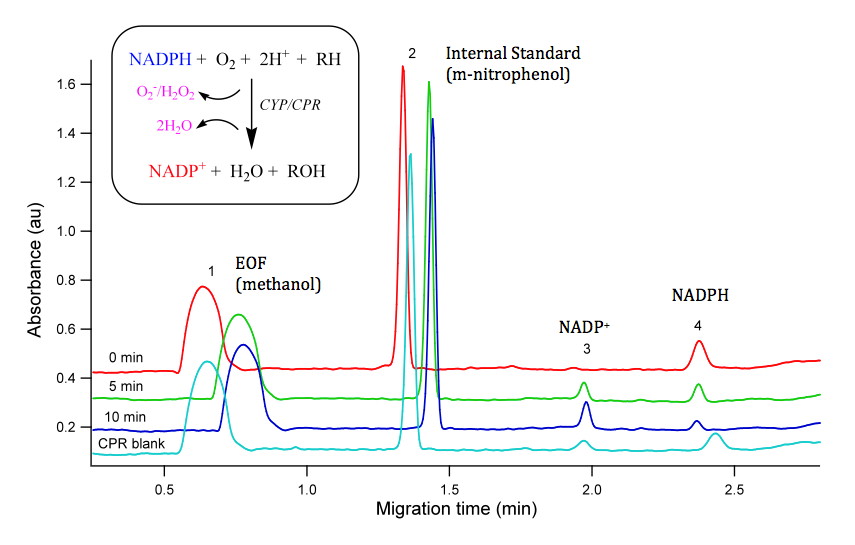
So how do we test function of novel enzymes? The first step is typically to express the gene to provide a pool of enzyme to work with. In our case, we typically express CYPs of interest in bacteria with the human co-enzyme, cytochrome P450 NADPH reductase. We have expressed several CYP genes from zebrafish from the CYP1 and CYP3 family.
The next major step is to perform some biochemistry: enzymatic reactions containing our expressed protein, NADPH cofactor, and possible substrates of interest. We have used a number of fluorogenic probe substrates (e.g. resorufin based compounds) to asses metabolism to a fluorogenic metabolite but because most of these probe substrates were developed for mammals, they are less informative for non-mammalian species. While most enzymatic assays monitor the loss of substrate or generation of metabolite, these assays are very specific for a single substrate or a single metabolite. The measurement of NADPH consumption is an indirect measure of CYP catalytic activity but provides a generic assay for CYP activity which allows for screening of CYP function with any compound of interest. When we couple this indirect assay with the power of robotics, we can undertake high throughput screening of chemical libraries.
Our current research is aimed at using high throughput screening approaches to better understand CYP function. For example, the zebrafish CYP1A and CYP3C1 enzymes have been screened with thousands of compounds, using NADPH consumption as the endpoint. These screens have identified ~100 potential substrates. Follow up tests on the hits provide additional data on the CYP-substrate interaction such as kinetic parameters and reactive oxygen species production. This research is completed in McMaster’s High Throughput Screening Laboratory using libraries of chemicals derived from natural products and off-label drugs and environmental chemicals.
This research has been funded by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery and Accelerator Programs, the Canadian Foundation of Innovation (CFI), Ontario Innovation Trust (OIT), and US Environmental Protection Agency.